Mycobacteria and phage |
|
 |
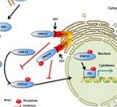
MycoMemSVM: Identification of mycobacterial membrane proteins and their types |
 |
MycoSec: Identification of mycobacterial secretory proteins |
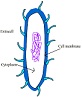 |
Mycosub: Predicting subcellular localization of mycobacterial proteins |
 |
PVPred: Identification of phage virion proteins |
 |
PHPred (version1.0) and (version 2.0): Prediction of bacteriophage proteins located in host cell |
 |
PHYPred: Prediction of bacteriophage enzymes and hydrolases |
 |
Lypred: Prediction of bacterial cell wall lyase |
Ion channel and toxin |
|
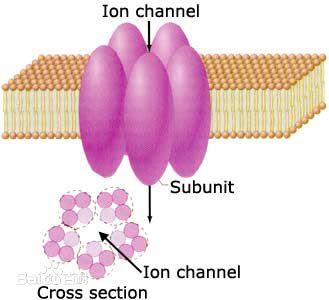 |
IonchanPred 2.0: Identification of ion channels and their types |
 |
iCTX-Type: Predicting the types of conotoxins in targeting ion channels |
 |
iVKC-OTC: Identifying the subfamilies of voltage-gated potassium channe |
Enzyme |
|
 |
ThermoPred: Identification of thermophilic proteins |
 |
AcalPred: Discriminating acidic enzymes from alkaline enzymes |
Sub-subcellular localization |
|
 |
ChloPred: Prediction of subchloroplast locations of proteins |
 |
TetraMito: Prediction of submitochondria locations of proteins |
 |
subGolgi: Prediction of subGolgi locations of proteins |
Anti-diseases |
|
 |
CaLecPred: Predicting the cancerlectins |
 |
IGPred: Identification of immunoglobulins |
 |
C2Pred: Prediction of cell-penetrating peptides |
 |
ApoliPred: Identification of apolipoproteins |
 |
HBPred and HBPred2.0: Identification of hormone-binding protein |
 |
TFPred: Predicting Preference of Transcription Factors for Methylated DNA |
 |
iBLP: Predicting Bioluminescent proteins |
Modification |
|
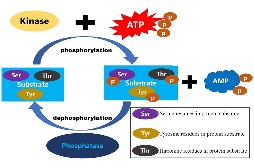 |
iPhoPred: Predicting the Phosphorylation Sites in Human Protein |
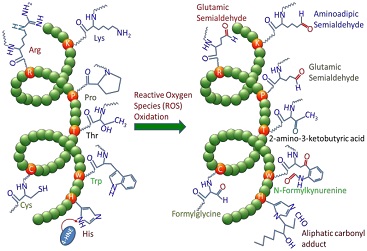 |
iCarPS: identifying protein carbonylation sites |
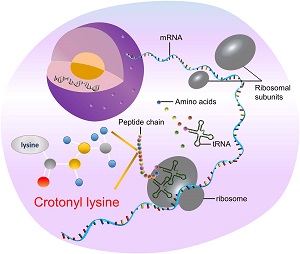 |
Deep-Kcr: detection of lysine crotonylation sites |
 |
iRice-MS: detection of lysine six modifications in rice |