You can visit the LinDing Group
to view more scientific achievements.
From this page, you may understand how to use our web server to start your prediction.
Tutorial
Question 1. How to make your prediction?
Answer: First, you should upload the protein sequences in fasta format. Secondly, you can choose a type of prediction in the 3-zone position of figure above. Finally, you can click the submit button in area 4 of the map and then wait for the result to appear.
Question 2. How to upload your own sequence?
Answer: There are two ways to upload a file. One way is to write the protien sequence with fasta format into the 2-zone position in the figure above, and you can view the sequence instance by clicking on the "example" option in area 4.
Question 3. What is the format requirements of fasta format sequence and file upload?
Answer:
Q1: What is fasta format sequence?
A1: A sequence in FASTA format begins with a single-line description, followed by lines of sequence data. The description line (defline) is distinguished from the sequence data by a greater-than (">") symbol at the beginning. To view the example sequence in FASTA format, please click the example button on area 4 of the figure above.
Q2: What is the request for the uploaded sequence file?
A2: First, the file must contain the fasta format sequence information. Second, the file suffix name must be ".txt", ".fasta" or ".fa".
Question 4. The detailed explanation of the page of prediction results
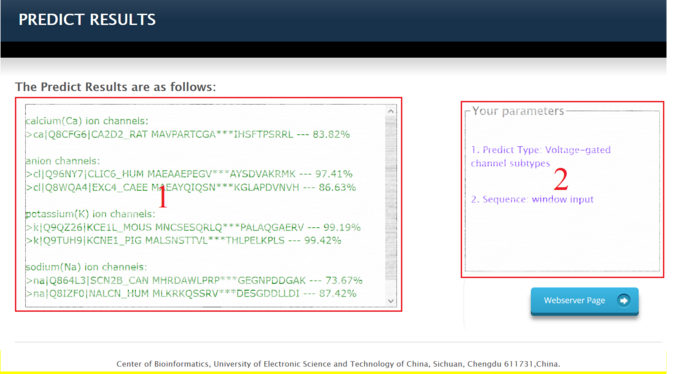

Answer: The prediction results interface consists of two parts. The area 1 in the figure above shows the predicted result information which including the prediction type of sequences, your protein sequences, and the
prediction
accuracy of the corresponding sequences. The area 2 shows the way you upload the sequences and the type of prediction information you choose.
That's all!
That's all!
